|
By: Terrance Lee
Investigators in the reproductive field seeking to use gene trapping encounter several difficulties. Gene symbols are somewhat variable and referencing them is an unreliable way to search the IGTC database; a more favorable way is to use a BLAST search of corresponding DNA sequences. Moreover, a substantial amount of time and effort is required to obtain a FASTA-formatted list of DNA sequences, perform a BLAST search on the IGTC database, and sort through the results to identify valid gene trap lines. Therefore, our group developed a program in Visual Basic named SpiderGene in order to minimize or eliminate these limitations, greatly facilitating our investigation. The user input of SpiderGene can be a spreadsheet list of gene symbols, GeneBank Accession numbers, or Unigene cluster numbers. The program will then proceed to search NCBI’s Unigene database, navigating through the Unigene cluster browser to the GenBank database and extracting the DNA sequence for the best EST sequence for each particular gene. This seemingly roundabout path also allows the user to use Unigene’s EST expression profile data to narrow down genes based on their expression profile if desired.
The sequences are then placed in FASTA format, which the user can use to BLAST the IGTC database. After results are generated, the user can paste the entire page of results into a window in SpiderGene, which will proceed to sort through the results based on a user-defined criterion of minimum bit score denoting a match. The end result is a list of genes fitting the criteria for a valid gene trap hit. While the advantages of the use of SpiderGene are primarily to save time and to decrease the amount of false negatives on the basis of using DNA sequences, there may be a number of false positives due to sequence similarity of genes of the same gene family or genes sharing a highly conserved protein motif. The number of false positives may become substantial when gene symbols are used as a result of unavoidable idiosyncrasies of NCBI’s search function. However, accuracy greatly increases with the use of GenBank Accession numbers or Unigene sequence cluster numbers. It should be noted that for most major centers, the sequencing and annotation process of trapped genes is an automated process. Therefore, recognizing the possibility of false positives, IGTC recommends that the researcher download the sequence tag and individual confirm the hit for correct annotation. Once the hits are confirmed, the user can follow the links provided on the IGTC site to order the cell lines from the research group.
SpiderGene was originally designed for in-house use but has been released for public use in the occasion that other researchers may benefit from its usage. Bugs have been assiduously minimized, but we regret that the program could possibly encounter unforeseen errors on different computers and is not compatible with Macs.
Download this installation folder (updated 1/30/07): SpiderGene.
Double-click the "setup" file and follow the instructions to install the program.
You may encounter some errors that you can ignore because the program will try to install out-of-date files or files that are in use already. These may include:
- Replace "RICHTX32.OCX" (RichTx32.ocx) with an older version? Click no.
- Replace "FM20.DLL" (Microsoft Forms DLL) with an older version? Click no.
- "msvcrt.dll" may be in use; Abort, Retry, or Ignore? Click ignore.
This is the menu:
 Option 1) "Search Unigene for Sequences" - allows the user to search the NCBI Unigene database for DNA sequences using gene symbols, GenBank Accession numbers, or Unigene sequence cluster numbers. The DNA sequences can be manually copied and pasted into the IGTC's search box for a BLAST search through their database. Option 1) "Search Unigene for Sequences" - allows the user to search the NCBI Unigene database for DNA sequences using gene symbols, GenBank Accession numbers, or Unigene sequence cluster numbers. The DNA sequences can be manually copied and pasted into the IGTC's search box for a BLAST search through their database.
Option 2) "Process Sequences on IGTC" - accepts the search results from the IGTC database and processes them quickly to identify gene trap cell lines that fit a minimum bit score supplied by the user.
Option 3) "Find Unique IGTC Hits" - accepts a list of hits from the IGTC search, which may have duplicates, and only returns the unique ones. This function can be expanded to take any list of genes or data and return only the unique ones, deleting the duplicates.
Option 4) "Process Expression Profile"' - accepts a list of genes and will search the NCBI Unigene database and returns genes that match user-defined criteria regarding their expression profiles.
OPTION 1: Search Unigene for Sequences
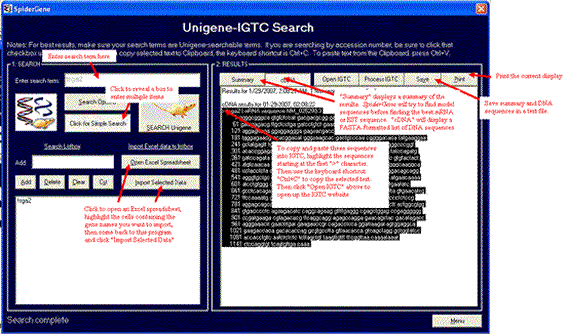
Enter your search terms via the single search box, the listbox ("advanced search"), or importing through an Excel spreadsheet. Then click the large Search button. Results are displayed on the right, which you can save or print. Highlight the FASTA-formatted DNA sequences and copy onto your Windows Clipboard using the keyboard shortcut Ctrl-C. Open up the IGTC website and paste these sequences into their BLAST search box.
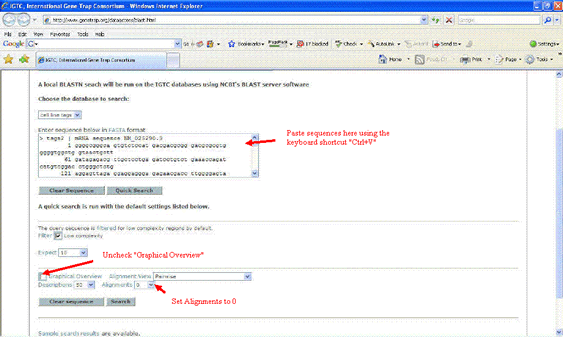
Paste DNA sequences onto IGTC and BLAST search them. (http://www.genetrap.org/dataaccess/blast.html)
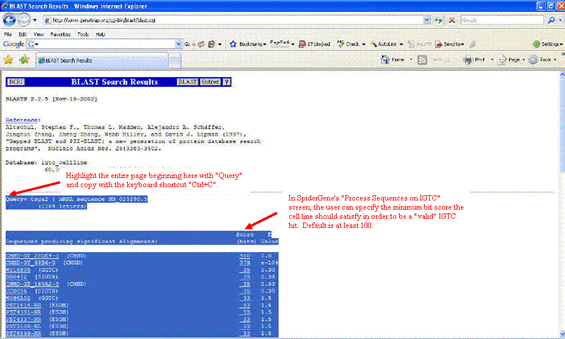
The IGTC results. Highlight everything beginning with the word "Query" and paste into Spidergene's "Process IGTC results" box.
OPTION 2: Process Sequences on IGTC
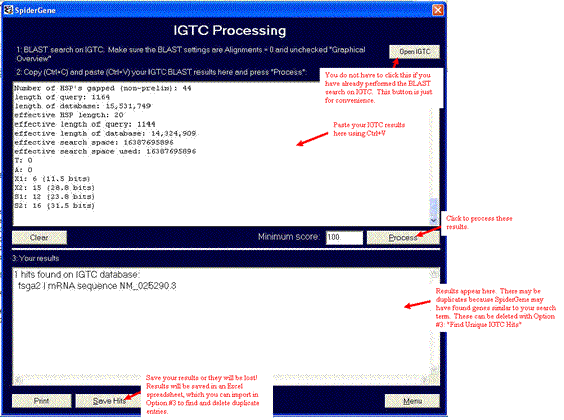
The IGTC results have been pasted into the first box using the keyboard shortcut for Paste, "Ctrl+V". Set the minimum bit score (default is 100) and press Process.
OPTION 3: Find Unique Hits
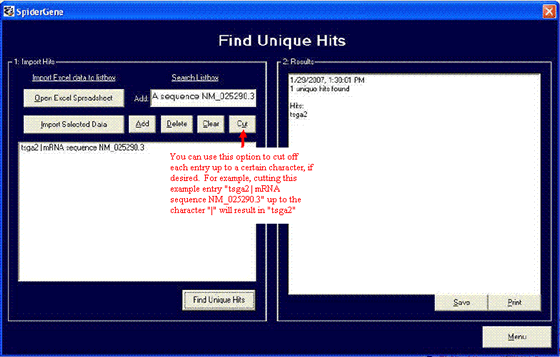
The format is similar to Option #1. Import the results you gathered in Option #2 after processing your IGTC results or enter them manually. Don't forget to save your results!
OPTION 4: Process Expression Profile
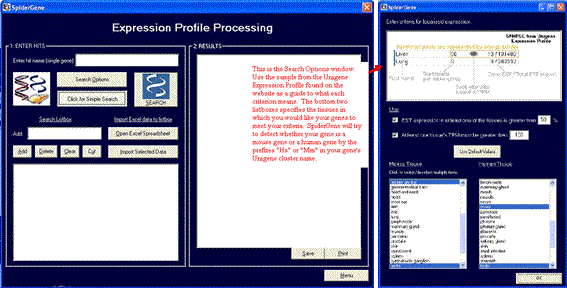
The format, again, is straightforward like Option #1. Enter a single gene or used advanced search to enter multiple genes manually or by importing from an Excel spreadsheet. Expression profile criteria is selected by clicking "Search Options".
|



