Presenting Author:
Bimal Chaudhari, M.D. M.P.H.
Principal Investigator:
Justin Starren, M.D.
Department:
Preventive Medicine
Keywords:
Genetics, Genomics, Informatics, ClinVar, Secondary Findings
Location:
Ryan Family Atrium, Robert H. Lurie Medical Research Center
C114 - Clinical
Variant Classification Conflict and Reporting of Secondary Findings
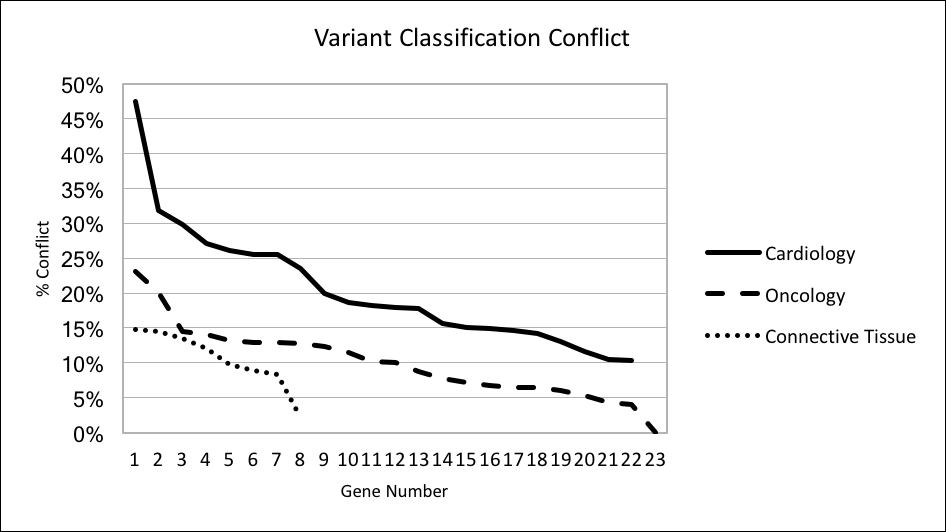
BACKGROUND: In clinical genomics, sequence variants are classified under standards put forth by the American College of Medical Genetics and Genomics (ACMG). When different labs classify the same variant differently a variant classification conflict (VCC) arises. This may lead to patients with the same underlying genetic risk being treated differently with consequences including under- and over-treatment. As the application of genomics in clinical settings increases, VCC is an increasingly recognized problem. The problem is exacerbated when secondary findings (SFs) are reported. The frequency of VCC has been described in specific genes and phenotypes (notably cardiomyopathy) but not for the generalizable case of genes subject to reporting of SFs under ACMG recommendations (ACMG56, 2015 version). OBJECTIVE: To calculate the frequency of VCC in the ACMG56. We hypothesize that the previously reported high rate of VCC in genes implicated in cardiomyopathy is not generalizable to all genes subject to reporting of SFs. METHODS: All submissions (N=136,210) of ACMG56 variants (N=39,876) in the ClinVar database were extracted. Submissions had their reported classification mapped onto the ACMG standard classification where able or were otherwise excluded (N=16,256). Variants with only a single submission were excluded from analysis (N=15,279). The percentage of eligible variants with conflicting variant classification was calculated for each gene. The genes in the ACMG56 were grouped into broad clinical categories by phenotype (Cardiology, Oncology, Connective Tissue, Other). Comparison was made between the Cardiology group and all other groups. RESULTS: There were 24,597 variants (107,697 submissions) eligible for analysis. The overall rate of VCC was 13%. The rate of VCC in specific genes ranged from 0% (NF2) to 47% (KCNQ1). The rate of VCC for Cardiology, Oncology, Connective Tissue and Other was 21%, 11%, 11%, 17% respectively (p<10^-5 for test of equality). Sub-group analysis dividing the Cardiology group between cardiomyopathy and non-cardiomyopathy genes did not effect results. DISCUSSION: The previously reported high rate of VCC in cardiomyopathy genes appears to also affect genes implicated in arrhythmia but is not universal across all genes recommended for reporting of SFs. Labs and healthcare systems reporting SFs should individualize their approach to surveillance for VCC based on particular genes and phenotypes.