Presenting Author:
Carlos Martinez, Ph.D.
Principal Investigator:
Panagiotis Ntziachristos, Ph.D.
Department:
Biochemistry and Molecular Genetics
Keywords:
topological domains, T-cell leukemia, Hi-C, Notch1 pathway
Location:
Third Floor, Feinberg Pavilion, Northwestern Memorial Hospital
B10 - Basic Science
Nuclear architecture changes during progression of acute leukemia
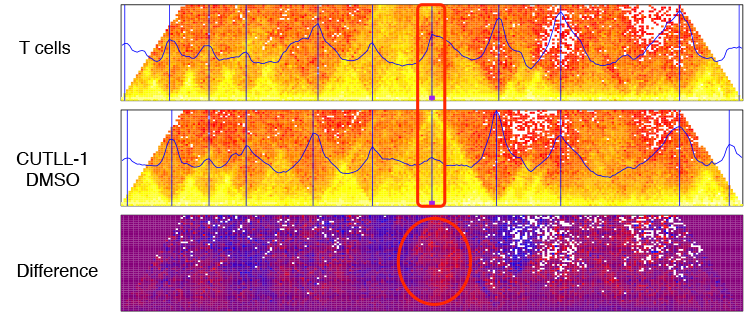
Mounting evidence suggests that the mammalian genome consists of mega-scale domains of highly interacting chromatin known as topological domains (TADs). We hypothesize that TAD boundaries are disrupted in T-cell leukemia leading to TAD merging, enhancer hijacking and aberrant gene activation. To test our hypothesis and ensure clinical relevance, we profiled primary patient samples using Hi-C and compared them to normal T cells. We developed a novel machine learning method for Hi-C data analysis, coupled with a novel method for TAD disruption detection. Extensive benchmarking of our methods using HiC-bench, our newly-developed benchmarking platform, demonstrated that we improve on well-established methods for TAD detection. Comparison of primary leukemia and normal T cell Hi-C datasets using our methodology, revealed TAD boundary disruptions near known oncogenes, and concomitant expansion of long-range promoter-enhancer interactions. Furthermore, Notch1 pathway inhibition led to the attenuation of such interactions, suggesting a central role of Notch1 binding in promoter-enhancer looping. Additionally, we detected previously uncharacterized TAD disruptions near genes that are not bona fide oncogenes, but contribute to carcinogenesis through non-oncogenic addiction. Finally, we integrated high-throughput RNA-seq, epigenome and copy number variation data to further characterize the detected TAD changes. In conclusion, our study may inform future therapeutic interventions by identifying novel targets linked to TAD boundary disruptions.