Presenting Author:
Xi Zhang, Ph.D.
Principal Investigator:
Hara Levy, M.D.
Department:
Pediatrics
Keywords:
Cystic Fibrosis, microRNA, Genetics, Parent-offspring trios, serum
Location:
Ryan Family Atrium, Robert H. Lurie Medical Research Center
C89 - Clinical Women's Health Research
Cystic Fibrosis Epigenome Roadmap Mapping, From Transcripts to microRNAs and More
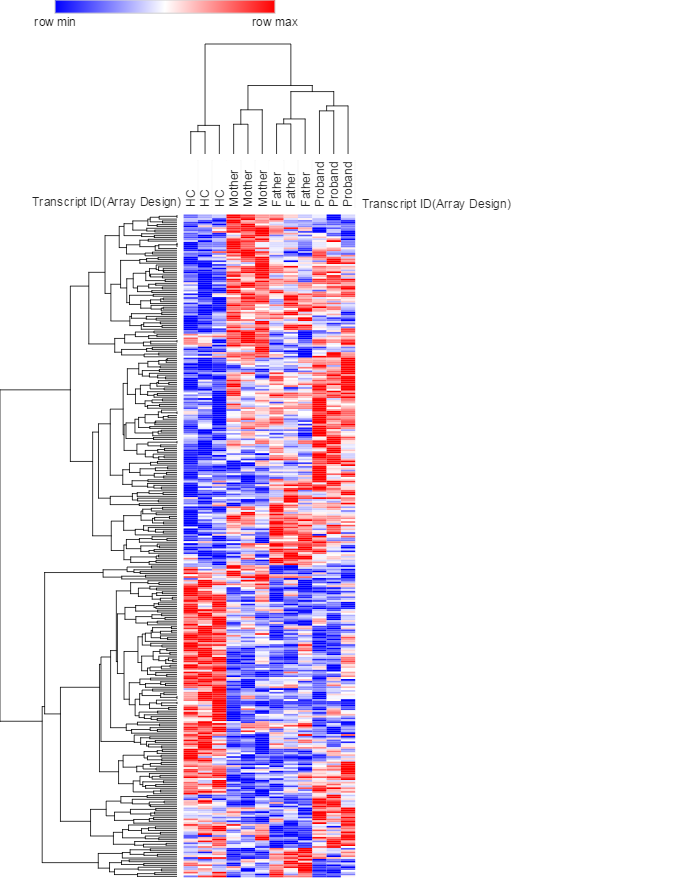
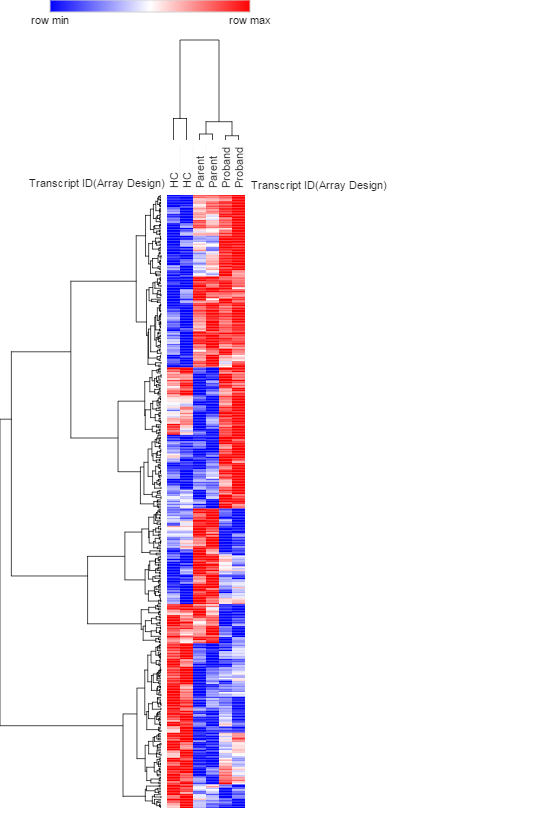
Background: The availability of the advanced genomic technologies for interrogating individual genomes offers unprecedented opportunities to apply genetics and epigenetics to medicine. We therefore combined two independent platforms (microarray and NanoString) to profile the miRNA (microRNA) expression and miRNA-mRNA networks in Cystic fibrosis (CF) associated sample. CF is a common lethal autosomal recessive Mendelian disorder that affects multiple organs including respiratory and digestive systems. CF patients normally inherited two copies of the defective gene (cystic fibrosis transmembrane conductance regulator, CFTR) from their parents who might just be healthy CF carriers. Over 1500 mutations of CFTR genes have been found in the newborns but it remains difficult to detect the correlation between CFTR genotype and clinical severity. Methods: This cohort study included over 20 selected CF probands (Class II) and their parents based on CFTR genotype. Clinical data on CFTR genotype, gender, sweat chloride level, airway pathogen, and pulmonary function were collected. Serum/plasma samples were collected and analyzed directly or used in cell treatments of peripheral blood mononuclear cell (PBMCs) models and THP-1 monocytes/macrophages cells involved in inflammatory response and cytokines production. Affymetrix genechip microarrays as well as NanoString nCounter technology were used to compare the expression difference of genes, miRNAs and whole-genome profiles in these samples. Results: Groups of genes coding pro-inflammatory cytokines were upregulated in CF plasma induced PBMCs (N=111, fold change > 2.0 or < -0.5, & adjusted p<0.05). NanoString analysis also indicated the innate immunity (e.g. macrophages) is activated in CF while adaptive immunity was impaired. miRNA microarray profiling revealed levels of several miRNAs, including precursor miRNAs and mature miRNAs were elevated (N=23, fold change > 2.0 or < -0.5, & adjusted p<0.10) in CF proband serum treated THP-1 monocytes, but not in healthy control (HC) group or parents groups. Measurement of miRNA expression showed the circulating miRNA profiles of CF proband and parents are similar and distinct from the HC samples. Conclusions and directions: This family based study enhances the understanding of unique miRNAs act and role as epigenetic modifiers and potential biomarkers that contribute in epigenetic reprogramming of the immune response across the CF generations. A next generation whole-genome microarray will be used to determine more details of the miRNAs profile during monocytes-to-macrophages activation. The relationship of the miRNA signatures and clinical variables including CFTR genotype and disease severity will be evaluated. The results will be tested through RT-PCR assays as well as a second group of subjects for validation.