Presenting Author:
Elizabeth Bartom, Ph.D.
Principal Investigator:
Elizabeth Bartom, Ph.D.
Department:
Biochemistry and Molecular Genetics
Keywords:
Bioinformatics, Genomics, NGS
Location:
Third Floor, Feinberg Pavilion, Northwestern Memorial Hospital
B3 - Basic Science
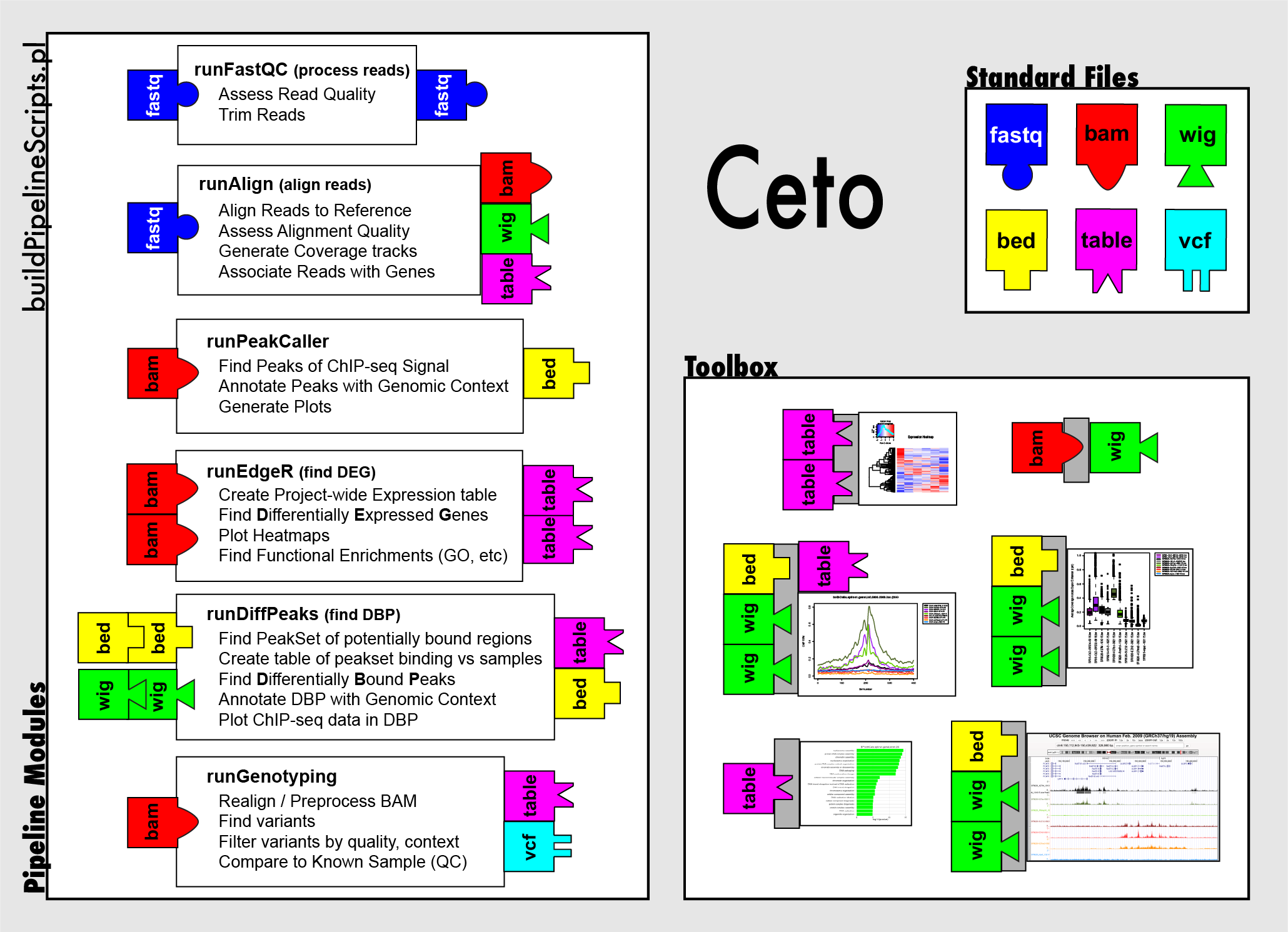
Ceto: A Flexible Framework for High Throughput Sequence Analysis
As the price of sequencing has dropped, the utility of sequence based assays has increased tremendously. Investigators can use immunoprecipitation-based methods like ChIP-seq to examine the genomic localization of DNA-binding proteins (from histones to transcription factors to RNA polymerase and anything inbetween) and sequence RNAs to determine the relative abundance of different genes and transcriptional isoforms. With these methods, we have the potential to gain unprecedented insight into the global workings of the cell. However, analyzing these large datasets remains a problem for traditional research labs. To address this need, I have developed a bioinformatics framework for sequence analysis, named Ceto. One part of Ceto consists of a large decision tree that generates analysis pipelines for RNA-seq and ChIP-seq datasets. These are set up to run on Northwestern's High Throughput Compute Cluster, Quest. Another part of Ceto is the "Toolbox", a set of modular R and perl scripts that can be run independently or incorporated into new analysis pipelines. Both parts of Ceto are available on GitHub at https://github.com/ebartom/NGSbartom .