Presenting Author:
Zachary Parton, B.A.
Principal Investigator:
Changiz Geula, Ph.D.
Department:
Cognitive Neurology and Alzheimer's Disease Center
Keywords:
Optical Clearing, iDISCO, CLARITY, Neurodegeneration, 3D modelling, Systems Neuroscience, Clinical Neuroscience, Clinica... [Read full text]
Location:
Third Floor, Feinberg Pavilion, Northwestern Memorial Hospital
B114 - Basic Science
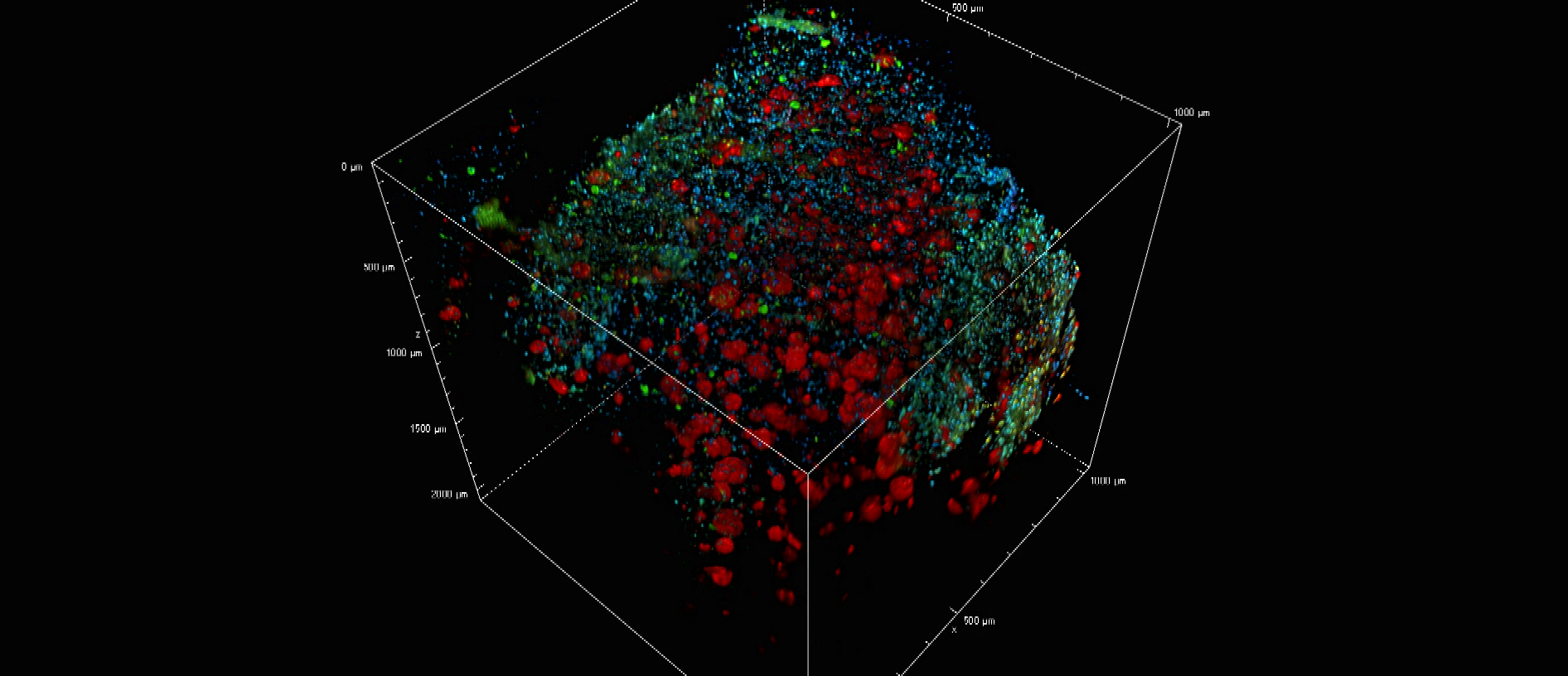
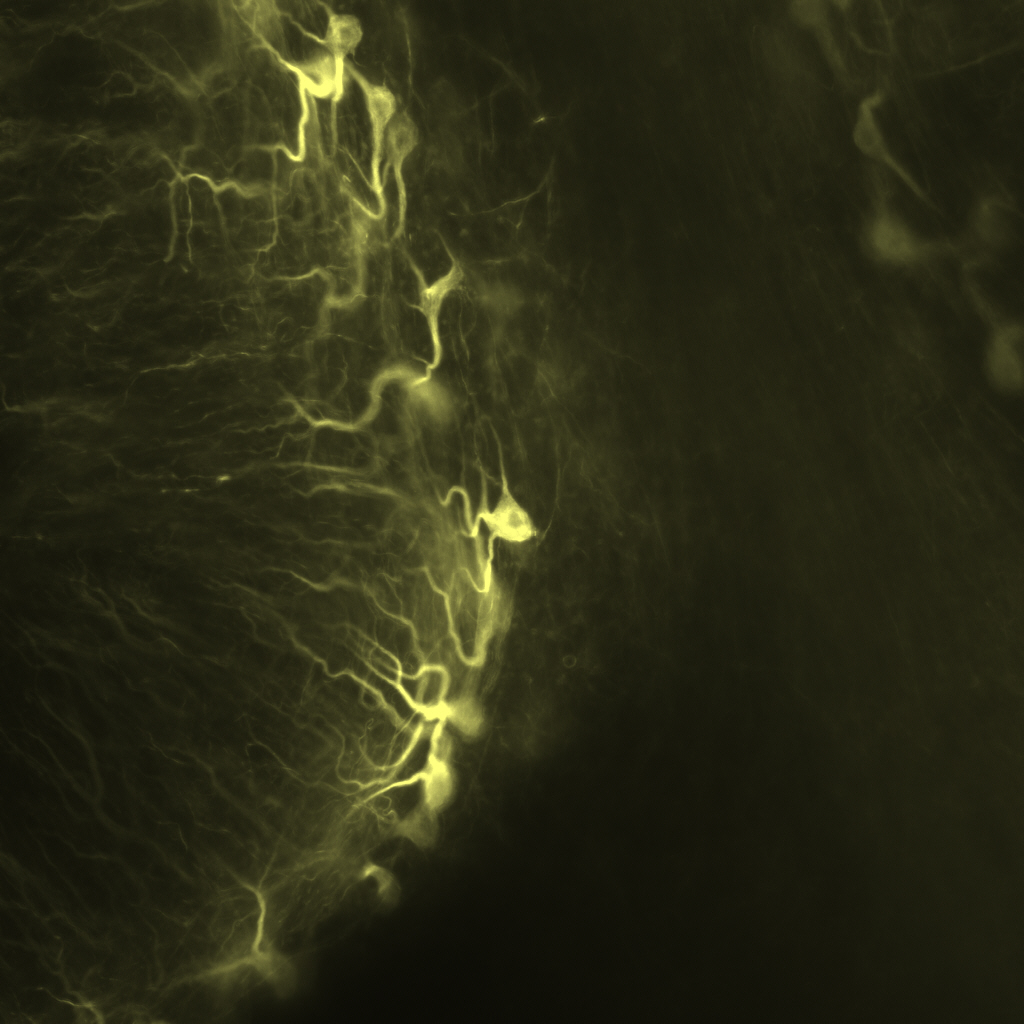
Clearing & 3D visualization of proteinomic markers in human brains
Recent advances in methods for optical tissue clearing have enabled 3D visualization of long-range neuronal circuitry and detailed cellular structures in a diverse range mammalian tissues. These procedures remove and/or homogenize lipid bilayers while preserving the structure of proteins, ultimately producing a transparent sample that can be fluorescently-labeled, and imaged. To date, few studies have specifically investigated the applications of these methods to specimens of human nervous tissue. The Cognitive Neurology and Alzheimer’s Disease Center (CNADC) at Northwestern University has worked to optimize clearing protocols for application to postmortem human tissue with the aim of conducting large scale 3D analysis of anatomic and pathologic features of neurodegenerative diseases. We have successfully applied the “CLARITY” and “iDISCO/+” protocols, and our trials have enabled us to fine-tune these procedures for relatively large human sections. We have validated labeling for neuronal, glial, and pathologic markers, including amyloid plaques, neurofibrillary tangles and TDP-43 inclusions. While both methods produced “cleared” cortical and cerebellar tissue, the solvent based iDISCO+ protocol has proved easy, reliable, cheap, and fast when compared with the relatively complicated and variable CLARITY protocol. We describe prototyped pipelines for study which generate high-resolution, high signal-to-noise 3-dimensional models (z-stacks) that can exceed 2mm, all while remaining comparable to classic IHC studies in terms of resources used. In sum, we present here methodologies which enable us to observe fine-scale anatomic distributions of pathology and directly inform systems-level understanding of intact and impaired cognitive functions.